Mutation Limited Evolution for Tandem Duplications
Duplications are a fundamental source of genetic novelty and genomic complexity and their prevalance in natural populations is expected to influence evolutionary trajectories. We have surveyed natural populations of D. yakuba and D. simulans to assess the number and types of gene duplicatons present among standing variation as well as the impacts that duplicates can have in adaptive evolution.
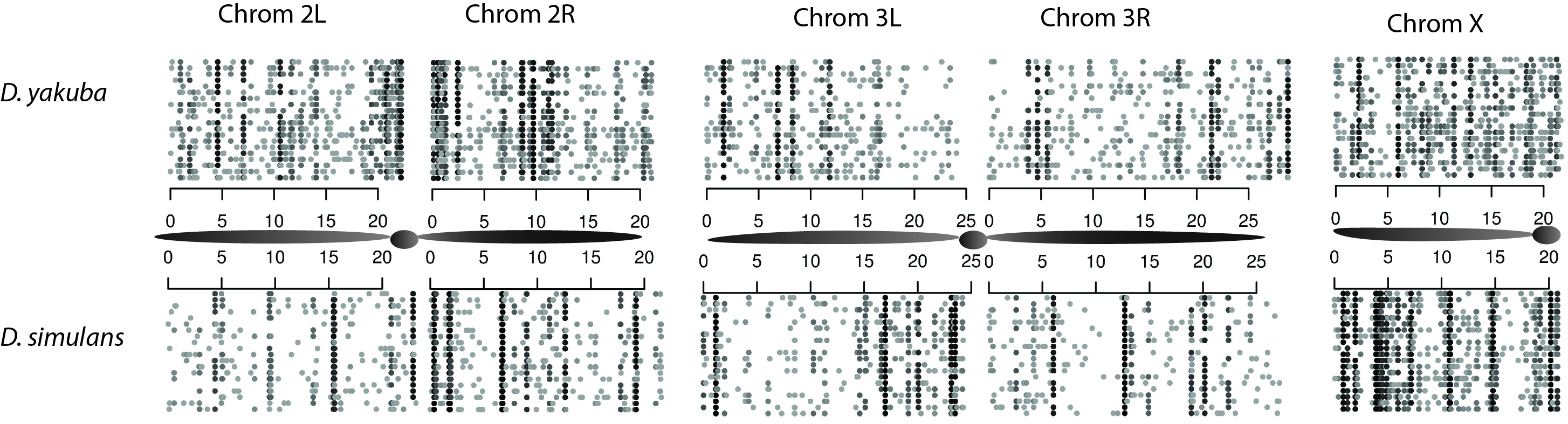 Figure 1. Observed variation in tandem duplications covers only a small percentage of the genome. Extrapolating to the entire population, we find that less than 15% of the genome will have a segregating duplicate in the population.
Figure 1. Observed variation in tandem duplications covers only a small percentage of the genome. Extrapolating to the entire population, we find that less than 15% of the genome will have a segregating duplicate in the population.
We have identified 1415 putative tandem duplications that are segregating in D. yakuba as well over 975 duplications in D. simulans. Throughout the entire population, we estimate that less than 15% of the genome is spanned by a segregating tandem duplication, suggeting that standing variation in populations is limited. We further estimate that mutation rates for whole gene duplication rates are low in comparison to SNPs, roughly 10-10-10-9, and mutation rates for chimeric genes and alternative structures are an order of magnitude lower.
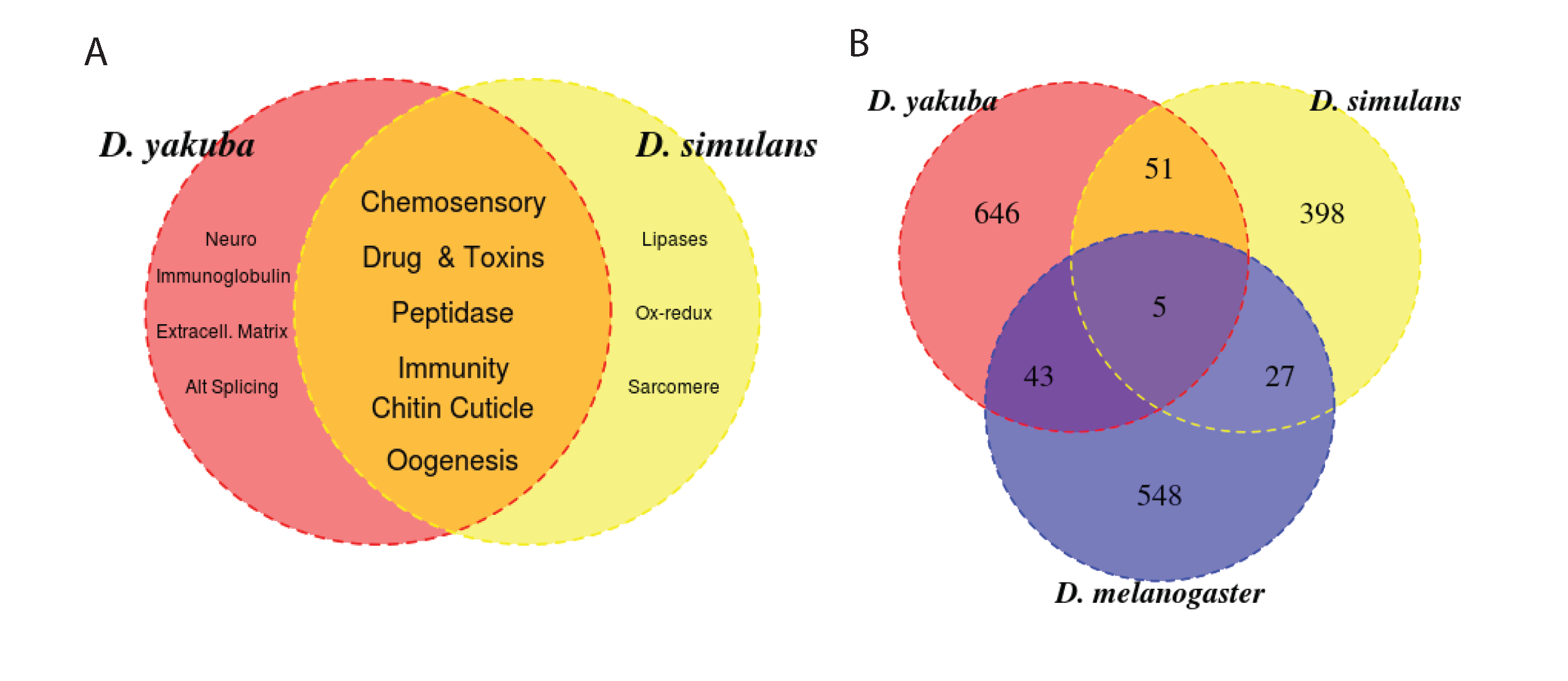 Figure 2. Comparing D. yakuba and D. simulans we observe strong concordance in the gene families that are overrepresented among tandem duplication, but very little agreement among duplicated genes.
Figure 2. Comparing D. yakuba and D. simulans we observe strong concordance in the gene families that are overrepresented among tandem duplication, but very little agreement among duplicated genes.
With low mutation rates and limited span of segregating variation, we find that the likelihood of adaptation from standing variation is 2-3% and that wait times for sweeps on new mutations are on the order of hundreds to thousands of years. Thus, if adaptation depends on specific tandem duplications, we expect that evolution will be severely limited by mutation. As a direct consequence of how mutation limits variation, we observe very few cases of parallel recruitment of tandem duplications for the same genes across different species. However, a strong concordance of functional classes across the genes that are duplicated points to a role for tandem duplications rapid evolution and Red Queen dynamics in both species.
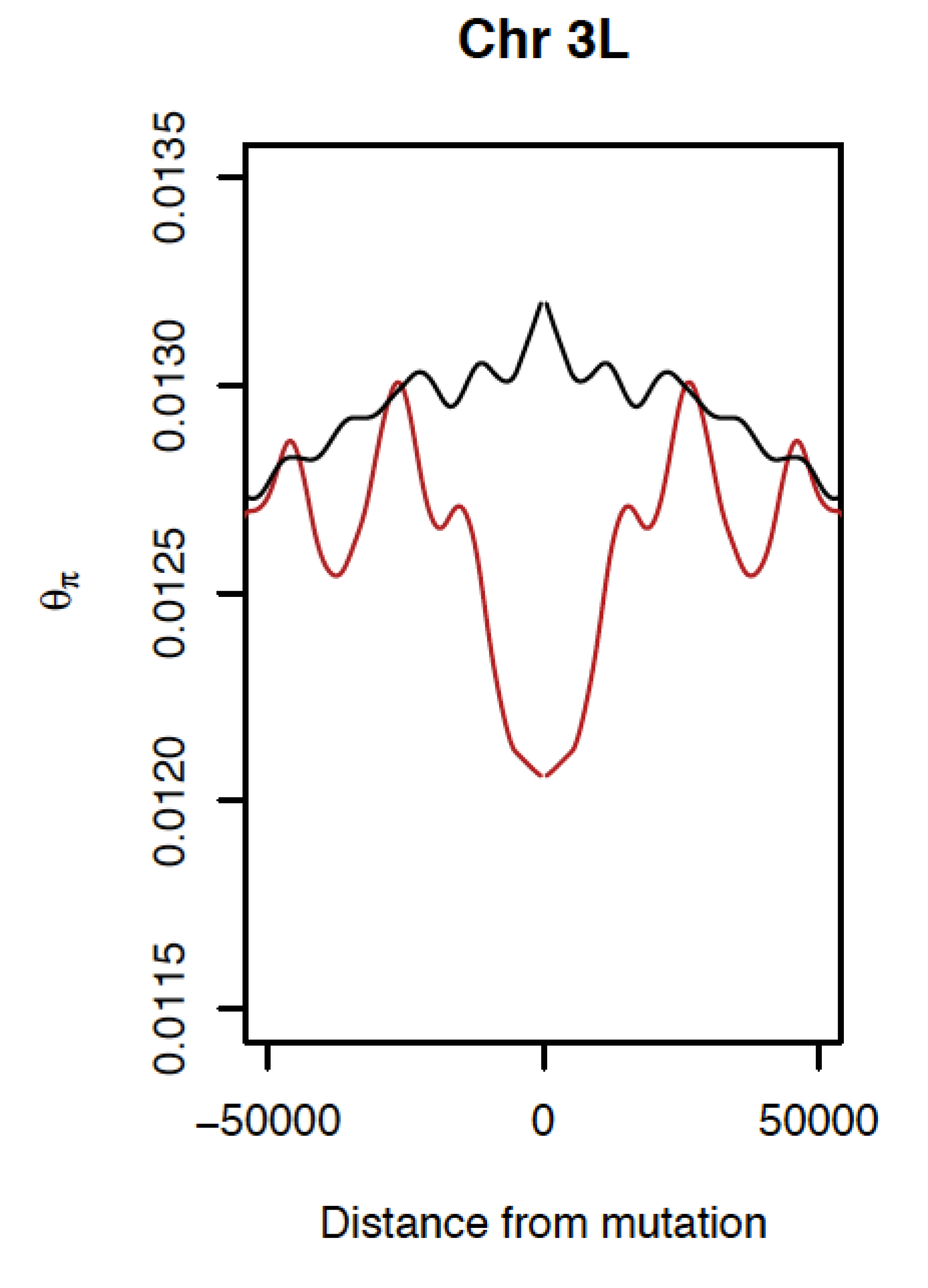 Figure 3. Reduced nucleotide diversity surrounding tandem duplications (red) compared to background for neutral intronic SNPs (black) on D. simulans 3L consistent with effects of selection.
Figure 3. Reduced nucleotide diversity surrounding tandem duplications (red) compared to background for neutral intronic SNPs (black) on D. simulans 3L consistent with effects of selection.
Selection
We observe an excess of tandem duplications on the D. simulans X chromosome in comparison with neutral intronic SNPs, suggesting widespread selection for tandem duplications on the X. We further observe signals of reduced nucleotide diversity in windows surrounding tandem duplicaitons on certain chromosomes, suggesting selective sweeps in progress. Thus, we suggest that tandem duplications are key players in adaptive evolution, especially with respect to rapidly evolving phenotypes.
Together, these results suggest that tandem duplications play an important role in evolutionary outcomes, but that limited substrates of standing variation and limited ability to generate new mutation will result in low levels of parallel duplication across species even with strong selective pressures for tandem duplicaitons. Thus, we suggest that suboptimal outcomes or alternative genetic solutions will be common during adaptive walks and that organisms will have limited ability to respond to rapid and drastic changes in environments.
