Tandem Duplications
Duplications are a fundamental source of genetic novelty and genomic complexity and their prevalance in natural populations is expected to influence evolutionary trajectories. We have surveyed natural populations of D. yakuba and D. simulans to assess the number and types of gene duplciatons present among standing variation as well as the impacts that duplicates can have in adaptive evolution.
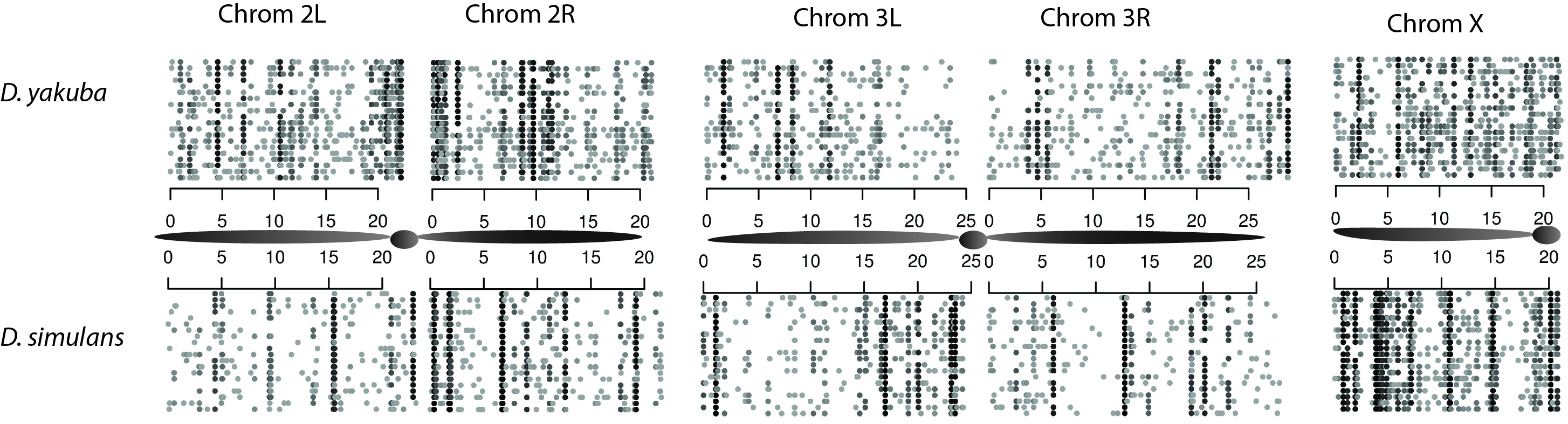 Figure 1. Locations of duplications identified in 20 strains of D. yakuba using paired end read mapping. Individual strains are represented by rows.
Figure 1. Locations of duplications identified in 20 strains of D. yakuba using paired end read mapping. Individual strains are represented by rows.
We have generated paired end Illumina sequencing reads for 20 strains of D. yakuba and 20 strains of D. simulans which we can use to identify tandem duplicationns in each species. Tandem duplications should manifest as reads that map in divergent orientation in contrast with properly paired reads.
We have identified 1415 putative tandem duplications that are segregating in D. yakuba as well over 975 duplications in D. simulans. These variants capture some 993 genes or gene fragments in D. yakuba and 711 genes or gene fragments in D. simulans. Among genes that are captured by tandem duplications, immune response, endopeptidases and lipases, drug and toxin metabolism, and chitin cuticle genes are overrepresented, suggesting that these variants play an important role in rapidly evolving phenotypes.
Complexity of variation
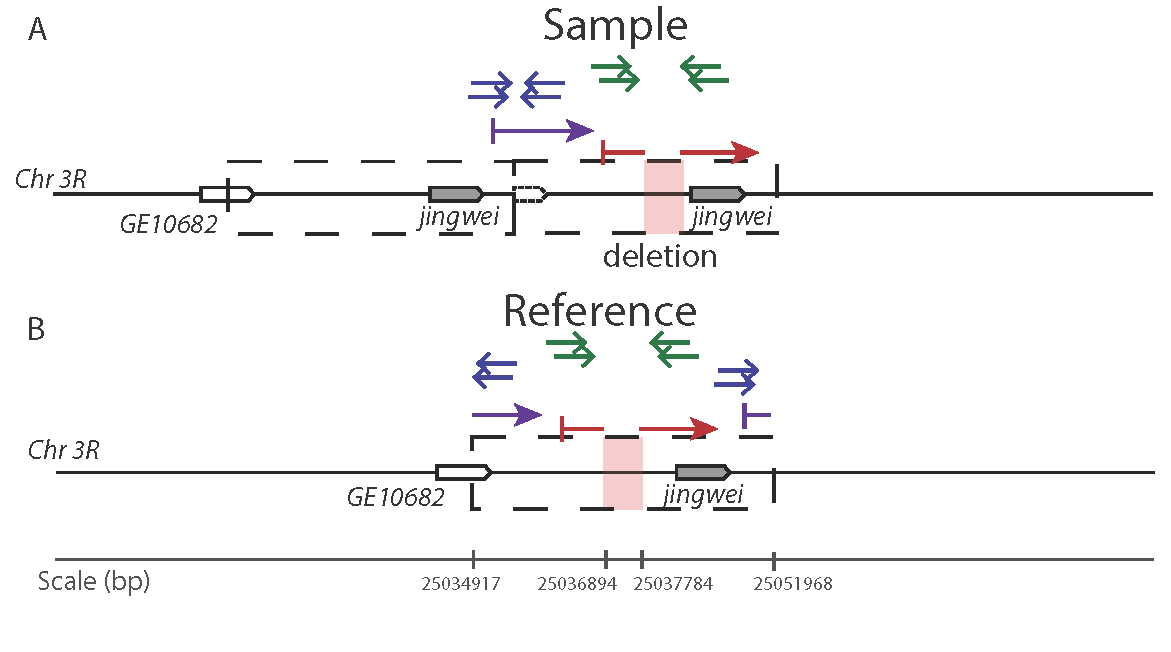 Figure 2. Read pairs identifying a duplication and subsequent deletion in the region surrounding the chimeric gene jingwei. A duplicaiton is indicated with divergenetly oriented read pairs (blue) as well as discontiguous alignment of PacBio long molecule sequencing (purple). A secondary deletion is present in a subset of duplicated alleles as indicated by long-spanning read pairs (green) and gapped alignment of PacBio sequences (red).
Figure 2. Read pairs identifying a duplication and subsequent deletion in the region surrounding the chimeric gene jingwei. A duplicaiton is indicated with divergenetly oriented read pairs (blue) as well as discontiguous alignment of PacBio long molecule sequencing (purple). A secondary deletion is present in a subset of duplicated alleles as indicated by long-spanning read pairs (green) and gapped alignment of PacBio sequences (red).
Many duplications show signs of secondary deletions in one or more copies of the region. Some 8% of duplications in D. simulans and 17% of duplications in D. yakuba display evidence of secondary deletions. The average deletion is 1.8 kb in D. simulans and 3.6 kb in D. yakuba. These deletions are larger than typical deletions in Drosophila but align well with the span of DNA that is typically excised via large loop mismatch repair (LLMR). The LLMR system is designed to remove excess unpaired DNA and has the potential to modify duplicated sequence quickly while alleles are still polymorphic, resulting in a richer source of genetic novelty than has been described in previous work on CNVs.
In D. yakuba, two independent duplications have captured the region surrounding the chimeric gene jingwei. One duplcate has experienced secondary deletiongs in one copy of the regions in a subset of strains. The secondary deletion lies just upstream of jgw resulting in more variation at this locus than is reflected through divergent read pairs alone.
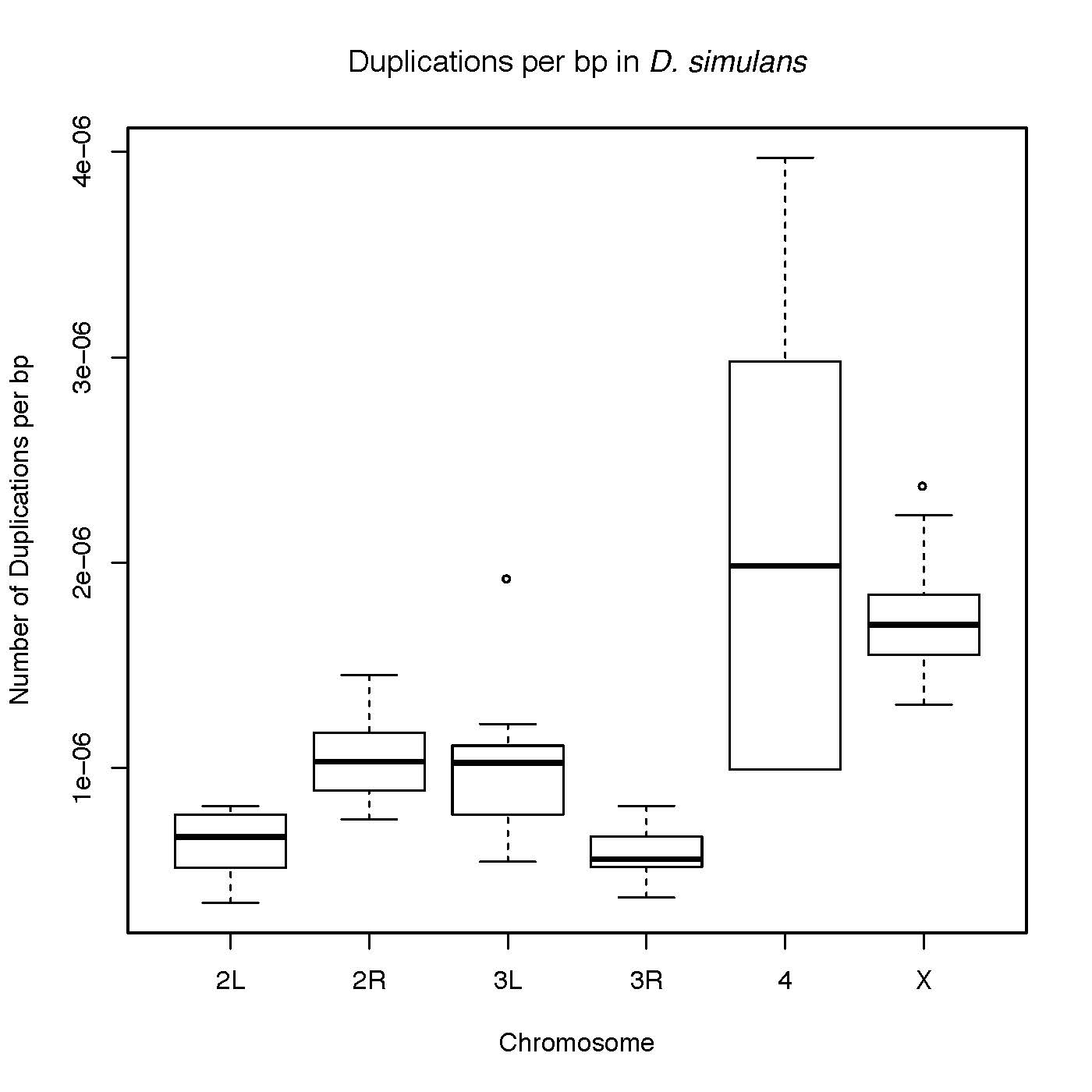 Figure 1. Locations of duplications identified in 20 strains of D. yakuba using paired end read mapping. Individual strains are represented by rows.
Figure 1. Locations of duplications identified in 20 strains of D. yakuba using paired end read mapping. Individual strains are represented by rows.
D. simulans X Chromosome
The D. simulans X chromosome contains an excess of duplicatons in comparison to each of the autosomes. The X chromosome also contains large numbers of high frequency variants, and large numbers of duplications that are flanked by repetitive sequence. These results likely speak to widespread selection on the X chromosome and repeated selective sweeps or demographic effects.
Chimeric Genes
We identify 78 chimeric genes in D. yakuba and 38 chimeric genes in D. simulans, as well as 143 cases of recruited non-coding sequence in D. yakuba and 96 in D. simulans . 10.4% of tandem duplications that capture genes in D. yakuba and 9.5% of tandem duplications that capture coding sequences in D. simulans form chimeric genes in agreement with rates of chimeric gene origination in D. melanogaster. Among parental genes in D. simulans, cytochromes and insecticide metabolism genes, sensory perception genes, and endopeptidase genes are overrepresented whereas cytochromes and toxin resistance are overrepresented in D. yakuba.
